Population structure of endangered spinetail devil ray (Mobula mobular) in the Lesser Sunda Seascape, Indonesia, revealed using microsatellite and mitochondrial DNA
November 2023
Muhammad Danie Al Malik, Mochamad Iqbal Herwata Putra, Edy Topan, Ni Luh Astria Yusmalinda, Ni Putu Dian Pertiwi, Yuliana Fitri Syamsuni, Ni Kadek Dita Cahyani, Enex Yuni Artiningsih, Sarah Lewis, Lumban Nauli Lumban Toruan, Muhammad Ghozaly Salim, Firmansyah Tawang, Faqih Akbar Alghozali, Derta Prabuning & Andrianus Sembiring
Keywords: Conservation genetics • Mobulids • Microsatellite DNA • mtDNA
Summary: The Lesser Sunda Seascape (LSS) hosts diverse megafauna like the spinetail devil ray (Mobula mobular). Drift gillnet fisheries have increased bycatch, endangering these species. To understand their genetic structure, 55 samples from East Flores, West Manggarai, and East Java were analysed using genetic markers. Results show two distinct subpopulations in the LSS: Savu Sea (East Flores and West Manggarai) and East Java. Despite this, coordinated conservation efforts in the LSS are crucial due to shared genetic traits among locations.
Abstract
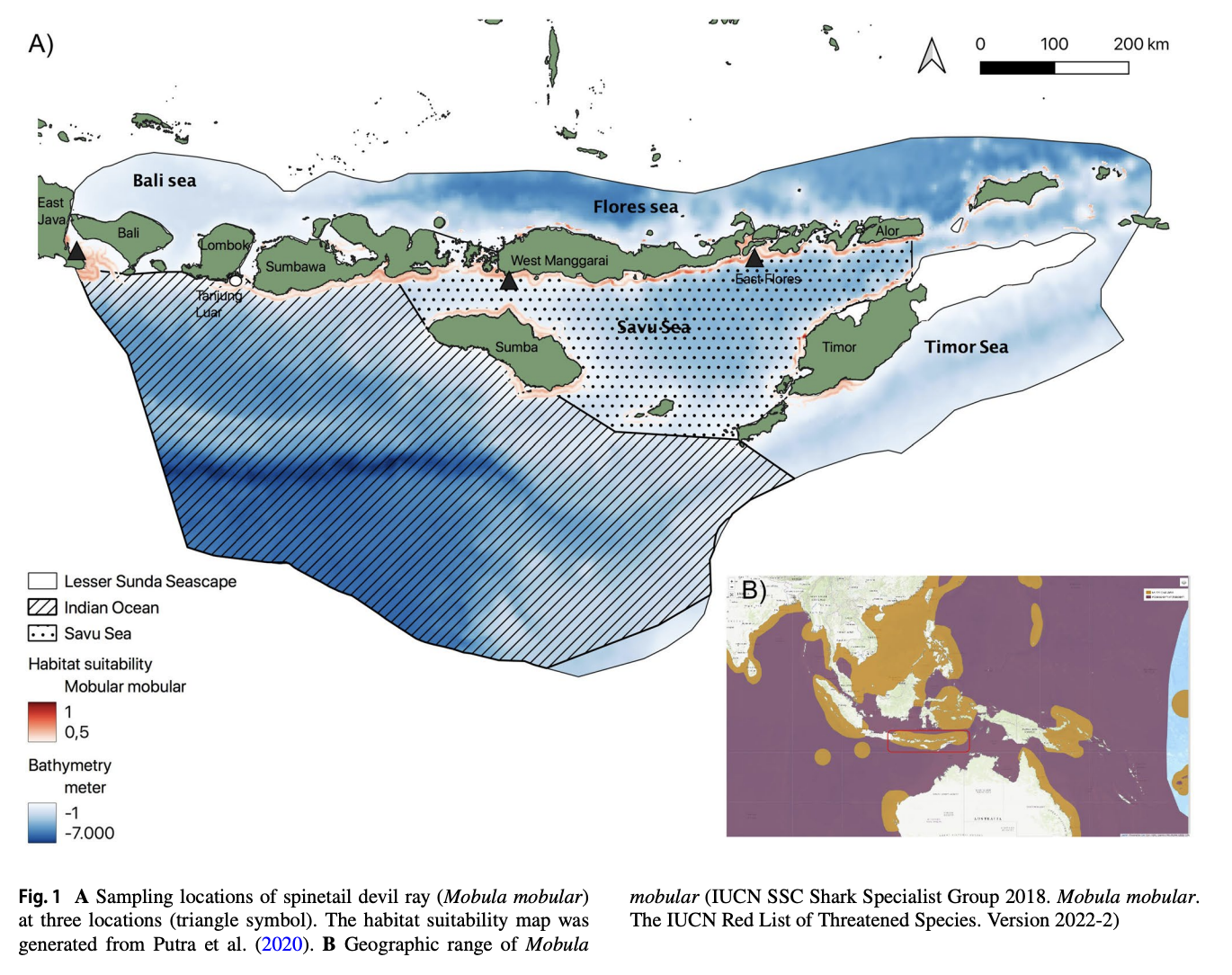
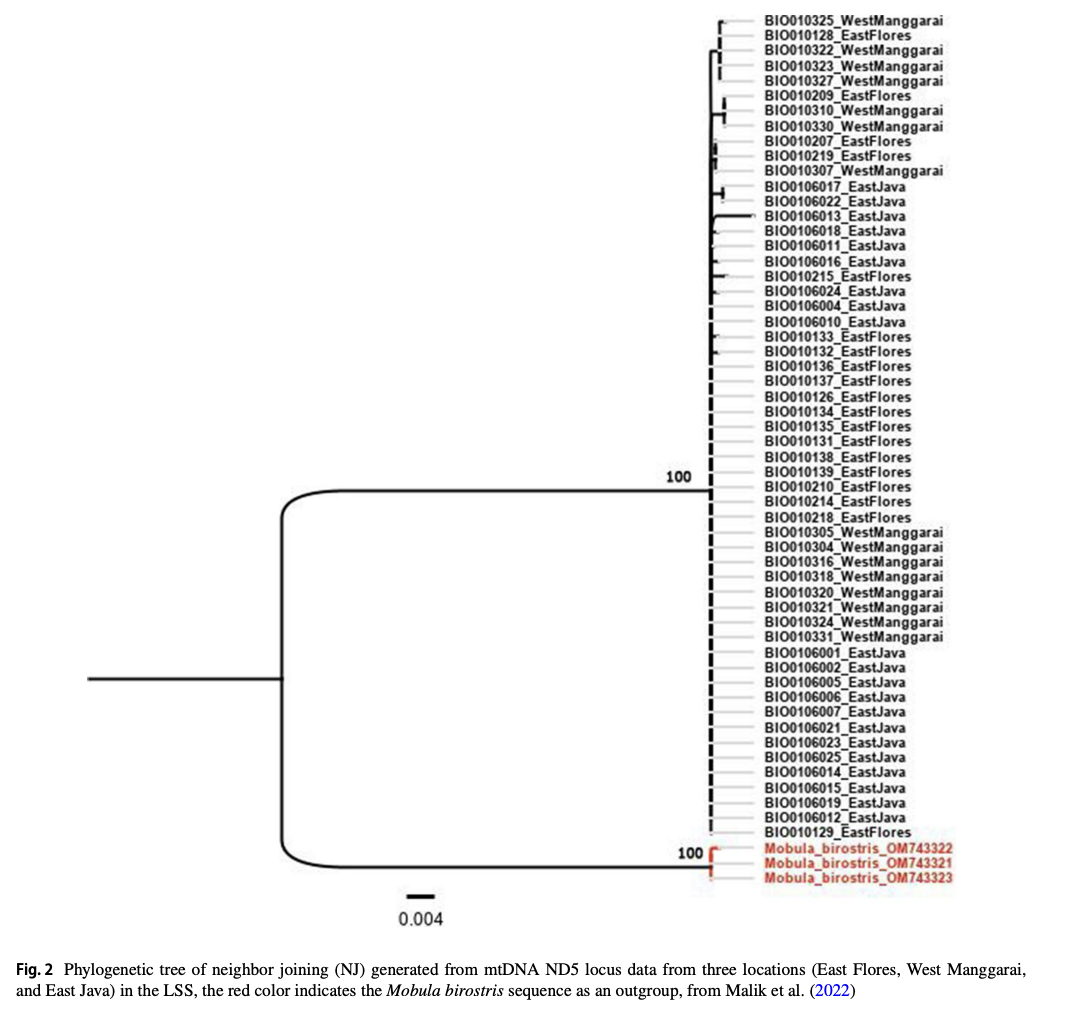
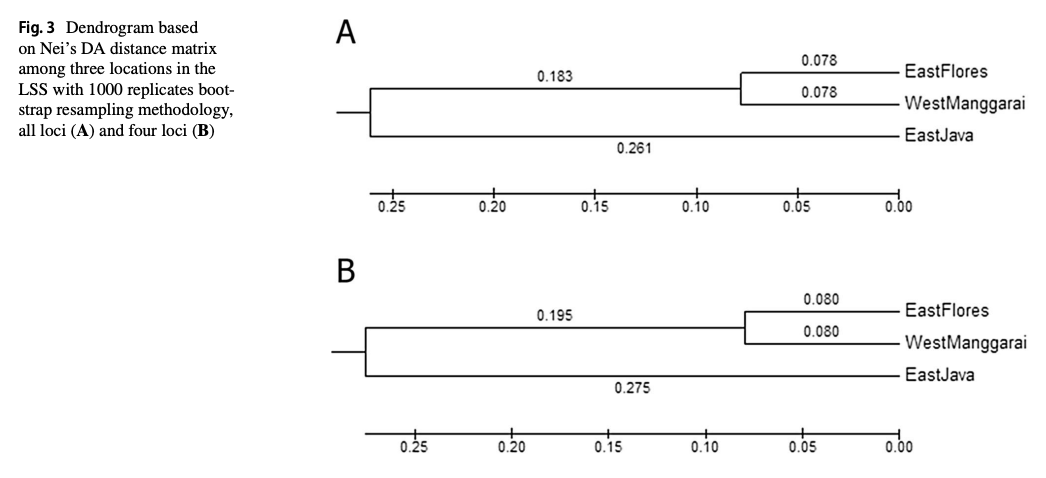
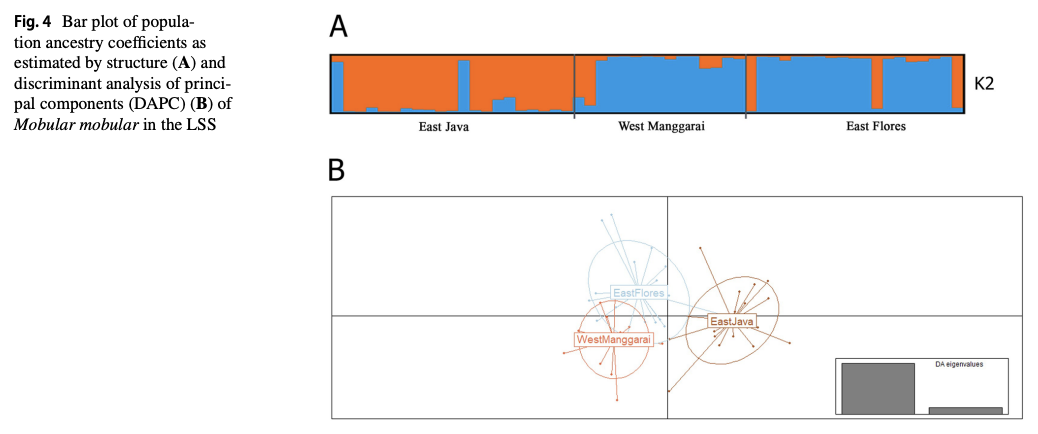
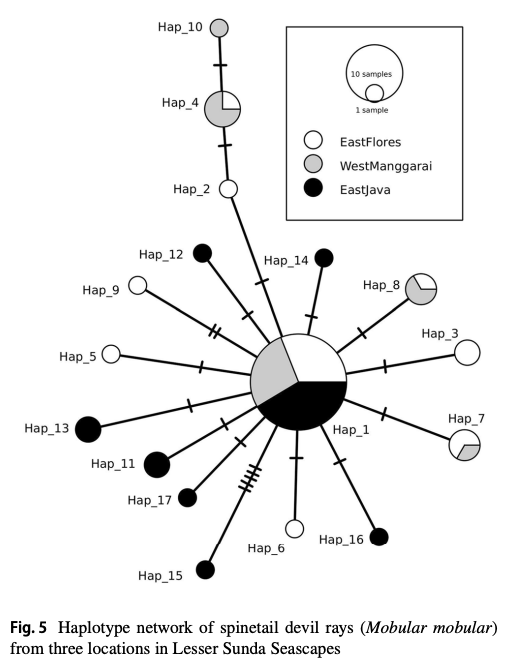
“The Lesser Sunda Seascape (LSS) is a critical habitat for various megafauna, including the spinetail devil ray (Mobula mobular). For decades, bycatch from drift gillnet fsheries has increased in the LSS, posing a greater threat to their populations. Migratory species require population-based management to efectively address their conservation needs and ensure their long-term survival. Fifty-fve samples were collected from three locations within the LSS, i.e., East Flores (19), West Manggarai (15), and East Java (21). This study aims to understand the population genetic structure of M. mobular within the three diferent locations around the LSS using eight microsatellite markers and mitochondrial DNA (mtDNA) NADH-dehydrogenase subunit 5. The analysis of molecular variances result indicated the signifcant result between the three locations based on microsatellite data (0.0876, p<0.05 for eight loci and 0.1360, p<0.05 for four loci) and mtDNA (0.04820, p<0.05). The pairwise genetic diferentiation index (Fst) revealed that East Flores versus West Manggarai has a nonsignifcant value (p>0.05). Meanwhile, East Flores or West Manggarai versus East Java showed a signifcant Fst value (p<0.05). In conclusion, microsatellite and mtDNA results revealed that in the LSS, spinetail devil rays are divided into two distinct subpopulations: the Savu Sea (East Flores and West Manggarai) subpopulation and the East Java subpopulation. Even though there are two potential subpopulations in the LSS, co-management of this species should be prioritized in the LSS due to the indication of shared haplotypes between those locations.”
Author Affiliations
Biodiversitas Indonesia (BIONESIA)
Department of Marine Science, Faculty of Fishery and Marine Science, Diponegoro University
Savu Sea Program, Misool Foundation
Focal Species Conservation Program, Konservasi Indonesia
Biology and Marine Fisheries Department, Faculty of Mathematics and Natural Science, Universitas Pendidikan
Marine ETP Species Program, Yayasan WWF Indonesia
Biology Department, Faculty of Science and Mathematics, Diponegoro University
Study Program of Aquatic Resource Management, Faculty of Fisheries and Marine Science, Nusa Cendana University
Mobula Project Indonesia (MPI)
Yayasan Hiu Pari Lestari (Elasmobranch Project Indonesia)
Funded by
Vulcan Inc.
MacArthur Foundation
Shark Conservation Fund
WildAid Canada Society
The Rufford Foundation
IDEA WILD
USAID
Pew Fellows Program in Marine Conservation at The Pew Charitable Trusts

