Phylogenomics and species delimitation for effective conservation of manta and devil rays
November 2020
Jane Hosegood, Emily Humble, Rob Ogden, Mark de Bruyn, Simon Creer, Guy M. W. Stevens, Mohammed Abudaya, Kim Bassos-Hull, Ramon Bonfil, Daniel Fernando, Andrew D. Foote, Helen Hipperson, Rima W. Jabado, Jennifer Kaden, Muhammad Moazzam, Lauren R Peel, Stephen Pollett, Alessandro Ponzo, Marloes Poortvliet, Jehad Salah, Helen Senn, Joshua D. Stewart, Sabine Wintner & Gary Carvalho
Keywords: ddRAD • Management • Mobulids • Single Nucleotide Polymorphism • Taxonomy
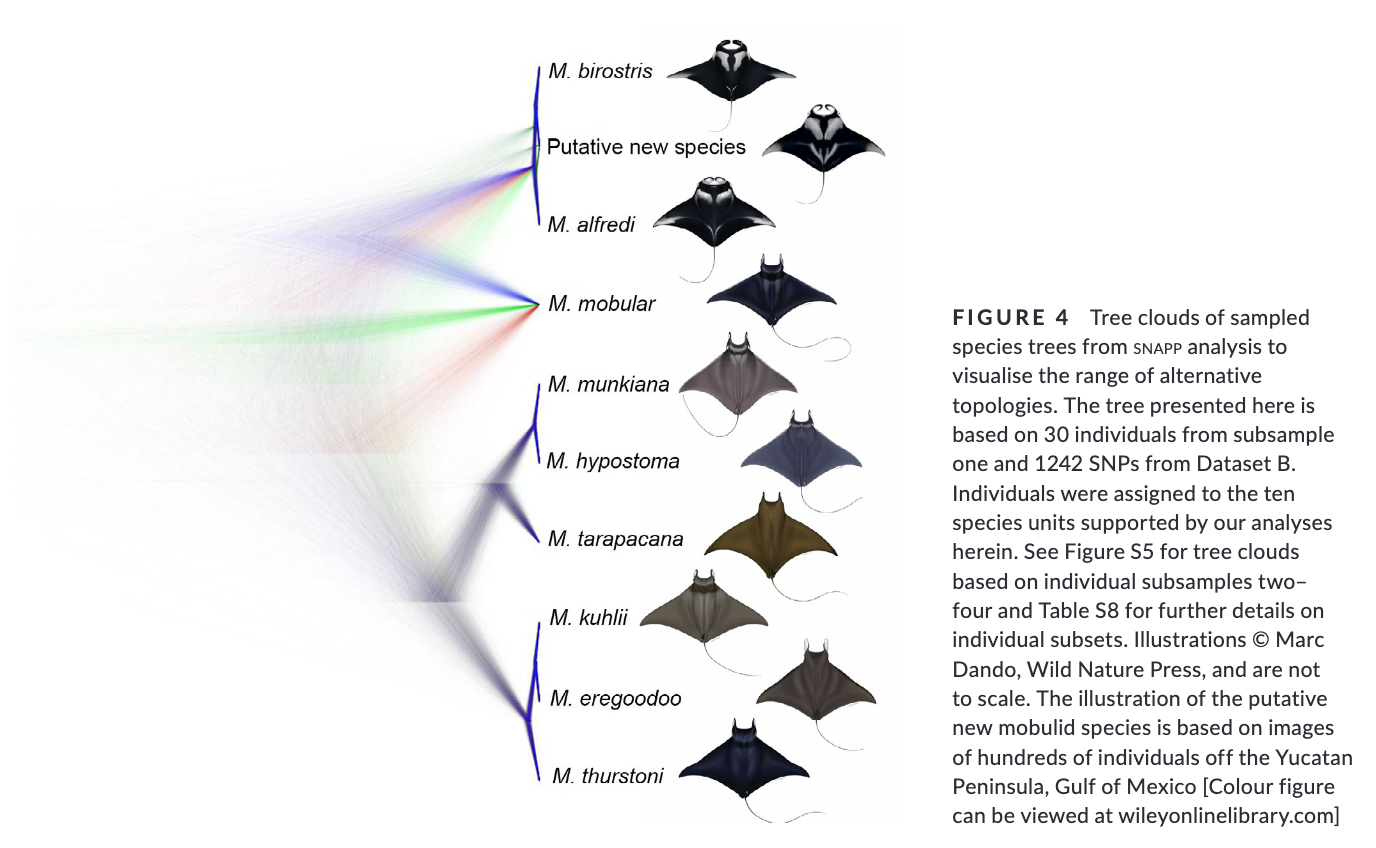
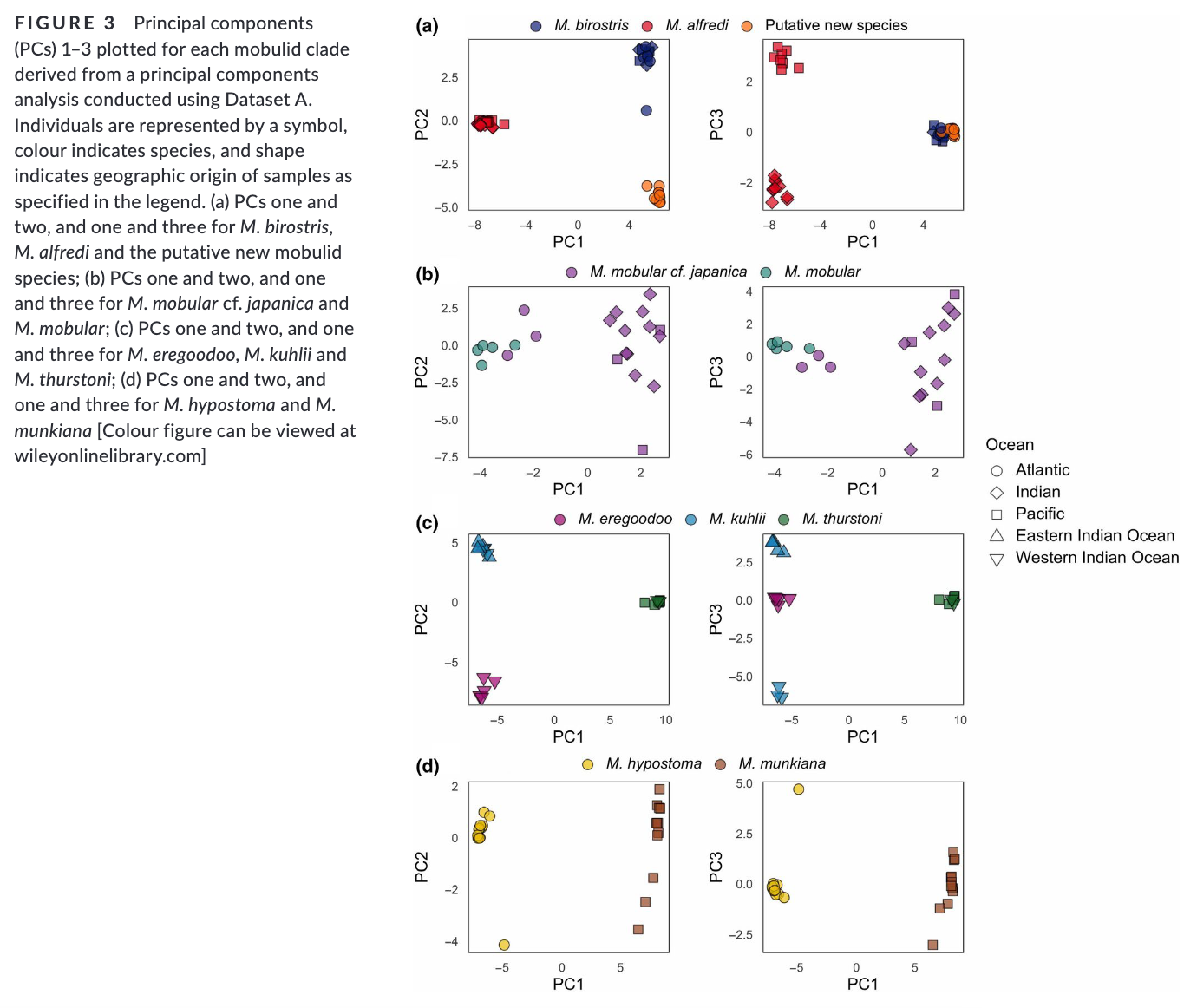
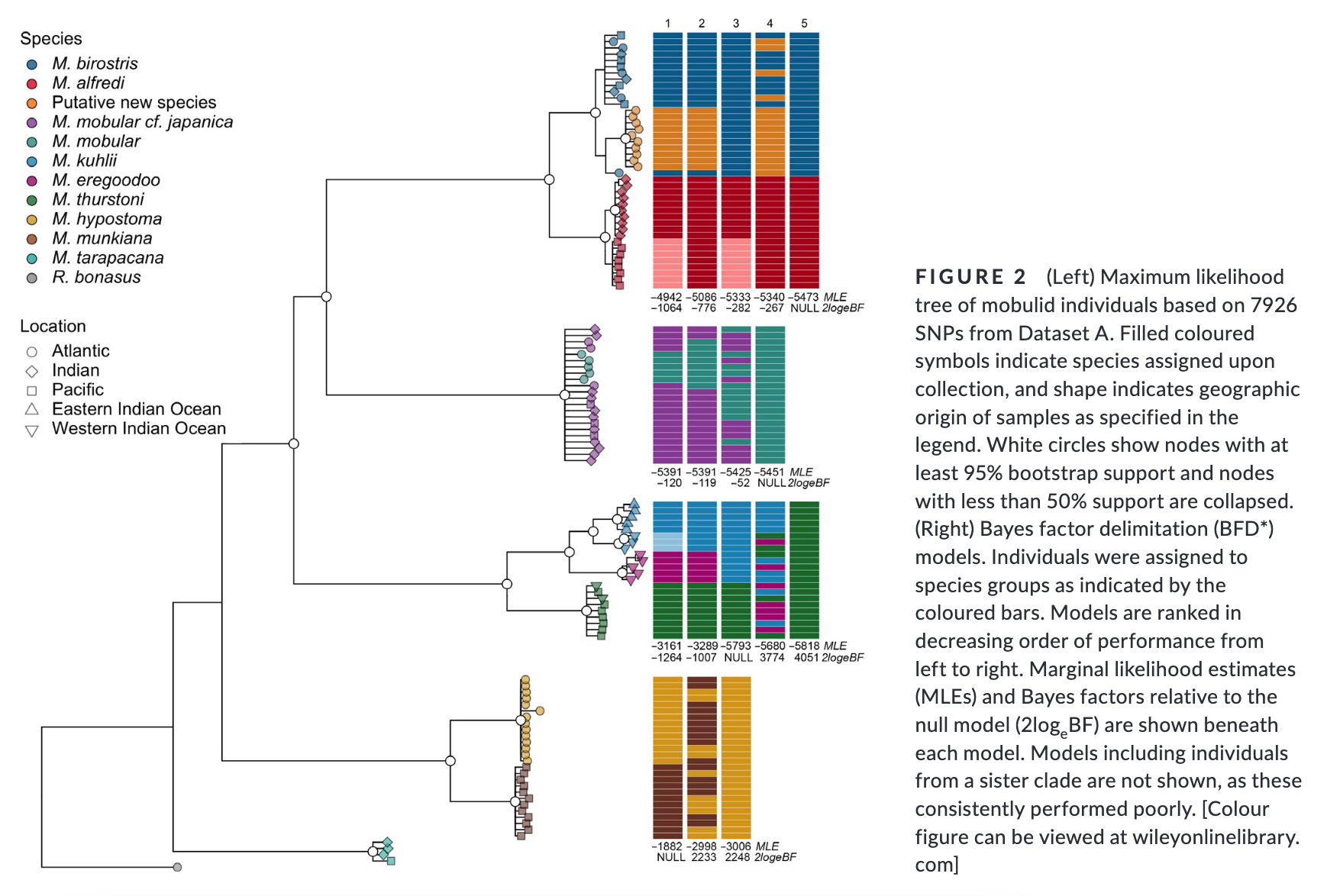
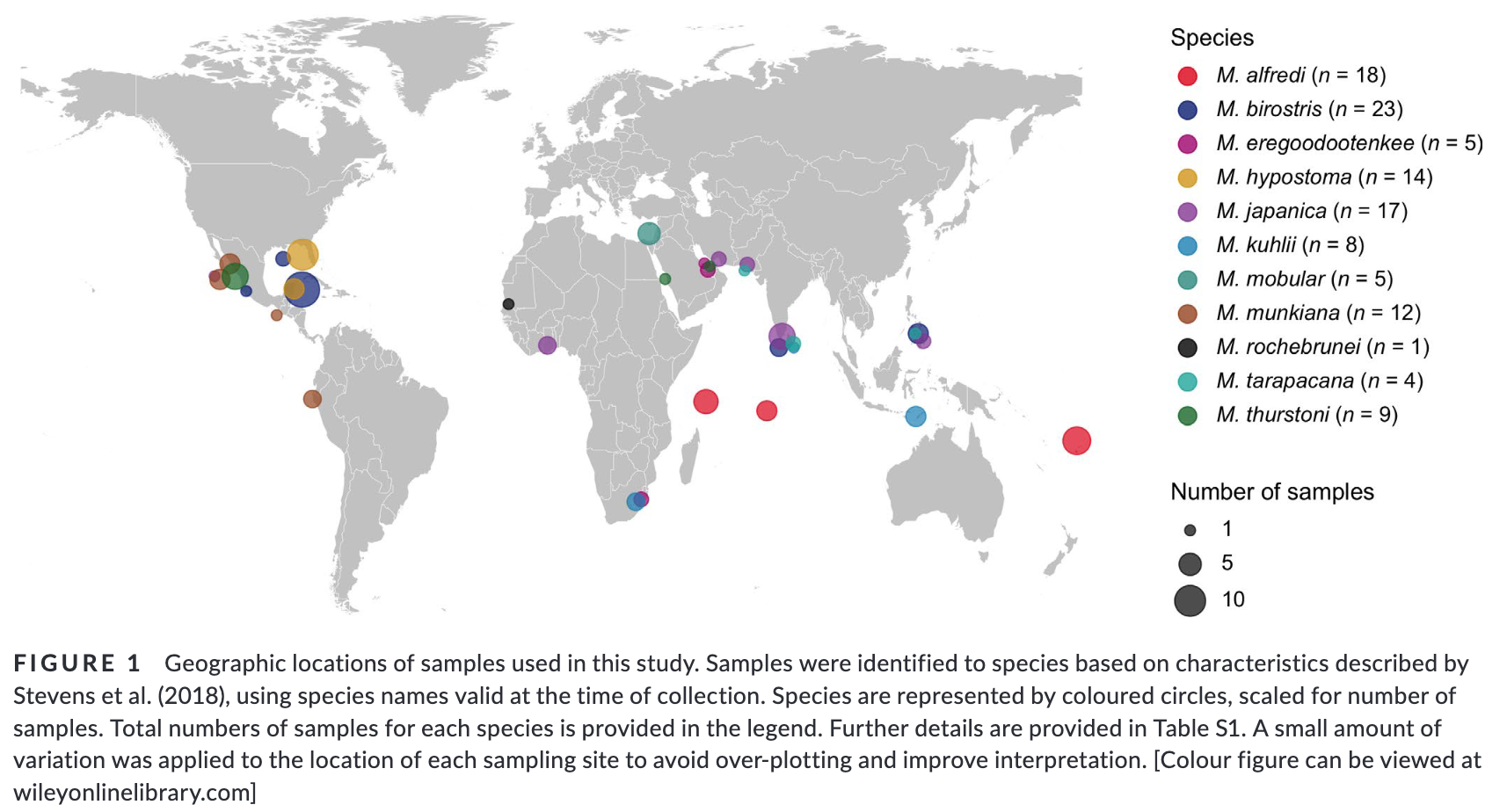
Summary: This study uses genome-wide SNP data to reconstruct the phylogenetic relationships and evaluate species boundaries, revealing independently evolving lineages and a potential new species in the Gulf of Mexico. Rapid speciation and incomplete lineage sorting have caused phylogenetic uncertainty. Cryptic diversity suggests management below the species level may be needed. The study provides a framework for species delimitation and highlights the potential of genomic data for conservation strategies.
Abstract
“Practical biodiversity conservation relies on delineation of biologically meaningful units. Manta and devil rays (Mobulidae) are threatened worldwide, yet morphological similarities and a succession of recent taxonomic changes impede the development of an effective conservation strategy. Here, we generate genome-wide single nucleotide polymorphism (SNP) data from a geographically and taxonomically representative set of manta and devil ray samples to reconstruct phylogenetic relationships and evaluate species boundaries under the general lineage concept. We show that nominal species units supported by alternative data sources constitute independently evolving lineages, and find robust evidence for a putative new species of manta ray in the Gulf of Mexico. Additionally, we uncover substantial incomplete lineage sorting indicating that rapid speciation together with standing variation in ancestral populations has driven phylogenetic uncertainty within Mobulidae. Finally, we detect cryptic diversity in geographically distinct populations, demonstrating that management below the species level may be warranted in certain species. Overall, our study provides a framework for molecular genetic species delimitation that is relevant to wide-ranging taxa of conservation concern, and highlights the potential for genomic data to support effective management, conservation and law enforcement strategies.”
Infographics
Author Affiliation
Molecular Ecology and Fisheries Genetics Laboratory, Bangor University
The Manta Trust
NERC Biomolecular Analysis Facility, Department of Animal and Plant Sciences, University of Sheffield
Royal (Dick) School of Veterinary Studies and the Roslin Institute, University of Edinburgh
TRACE Wildlife Forensics Network
School of Life and Environmental Sciences, The University of Sydney
National Research Center, Gaza City, Palestine
Mote Marine Laboratory, The Center for Shark Research
Océanos Vivientes A. C.
Department of Biology and Environmental Science, Linnaeus University
Blue Resources Trust
Department of Natural History, Norwegian University of Science and Technology (NTNU)
Elasmo Project
RZSS WildGenes Lab, Royal Zoological Society of Scotland
WWF Pakistan
School of Biological Sciences, University of Western Australia
The Australian Institute of Marine Science
Save Our Seas Foundation
Large Marine Vertebrates Research Institute Philippines
Tolheksbos 57
Ministry of Agriculture Directorate General of Fisheries, Gaza City, Palestine
KwaZulu-Natal Sharks Board
School of Life Sciences, University of KwaZulu-Natal
Funded by
Natural Environment Research Council
Marine Conservation Action Fund by New England Aquarium
Fisheries Society of the British Isles
Mote Scientific Foundation
People's Trust for Endangered Species
National Geographic Society
Genetics Society
Disney Conservation Foundation
National Marine Sanctuary Foundation
Save Our Seas Foundation
Marie Skłodowska-Curie



