Genomic Tools for Conservation and Management of Manta and Devil Rays (Mobula Spp.)
2015 - 2019
Dr. Jane Hosegood
Keywords: Manta • Mobula • Devil Ray • Elasmobranch • Management • Genetic Diversity • Population Dynamics • Single Nucleotide Polymorphism (SNP)


Summary: To effectively conserve biodiversity, a better understanding of genetic diversity and population dynamics is needed. The overexploitation of manta and devil rays through international trade puts these species at risk, leading to unsustainable fishing practices. Despite international regulatory frameworks to protect these species, knowledge gaps persist, including a lack of representative global tissue samples and limited understanding of stock structure, gene flow, and population dynamics. This study presents genome-wide Single Nucleotide Polymorphism (SNP) data as an approach to fill knowledge gaps. This study highlights mismatches between currently recognised species, reveals geographically mediated population structure in several species, and identifies standing variation in extinct ancestral populations as a driver of phylogenetic uncertainty. This study also raises concerns about peripatric speciation in isolation, anthropogenic climate change, and accompanying sea level changes and provides insights on population structure and adaptive divergence of related species of conservation concern.
Abstract
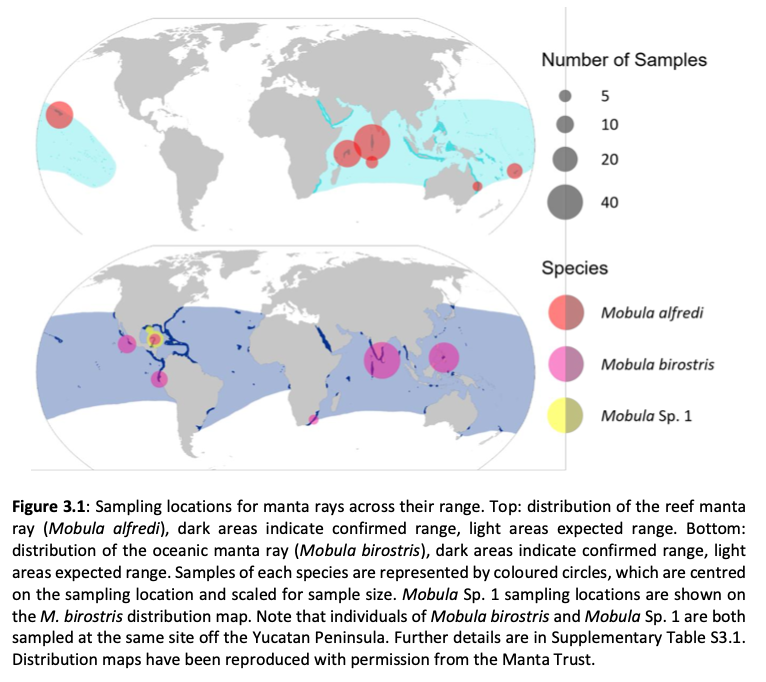
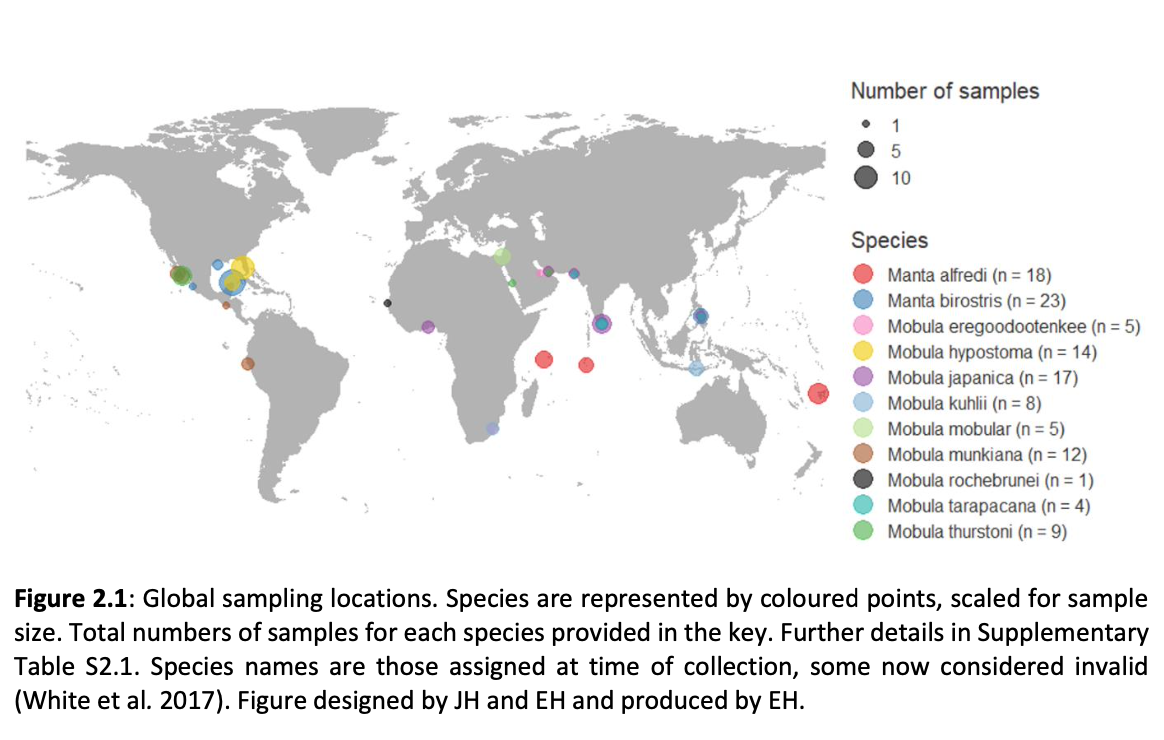
“Effective measures for the conservation of biodiversity require an understanding of the extent and distribution of diversity within and among species. Studies focussed on providing such understanding can benefit from increasingly powerful and accessible genome-wide data. Overexploitation of marine fisheries is a global problem resulting in loss of genetic diversity and declines in many species, and there is increased awareness and uptake of genomic principles in fisheries management and conservation. Manta and devil rays (Mobula spp.) are increasingly threatened by targeted and bycatch fisheries supplying the international demand for their gill plates. Such impacts will likely be met with catastrophic declines exacerbated by their slow life history traits, rendering these fisheries unsustainable. To alleviate threats, all species of mobulid ray are listed on CITES Appendix II to regulate international trade, and on CMS Appendices I and II to coordinate protection and implement conservation efforts. However, the status of mobulid rays is not matched by understanding of stock structure, gene flow, population dynamics, processes driving variation between species, and species boundaries. To date, a lack of representative global tissue samples, ongoing taxonomic ambiguity and ineffectual traceability measures combine to constrain the development and implementation of a coherent and enforceable conservation strategy for these species.
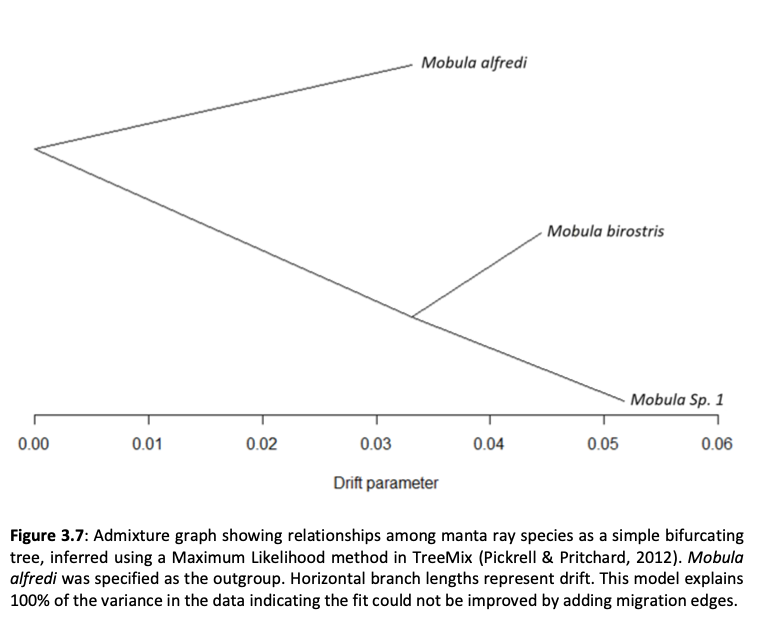
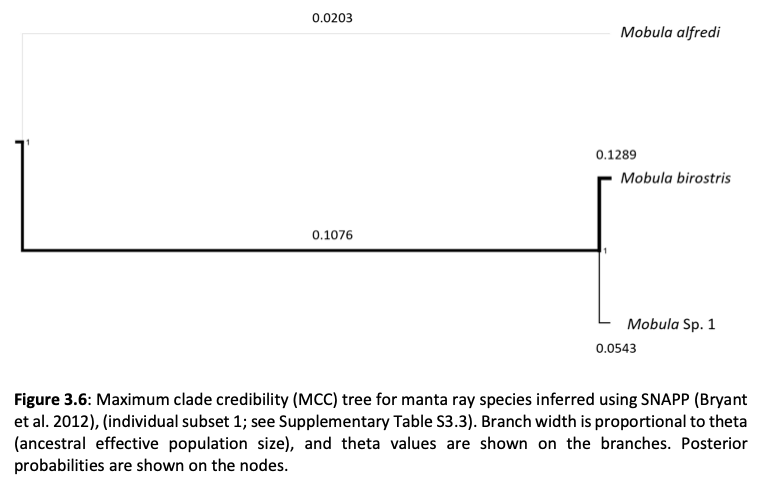
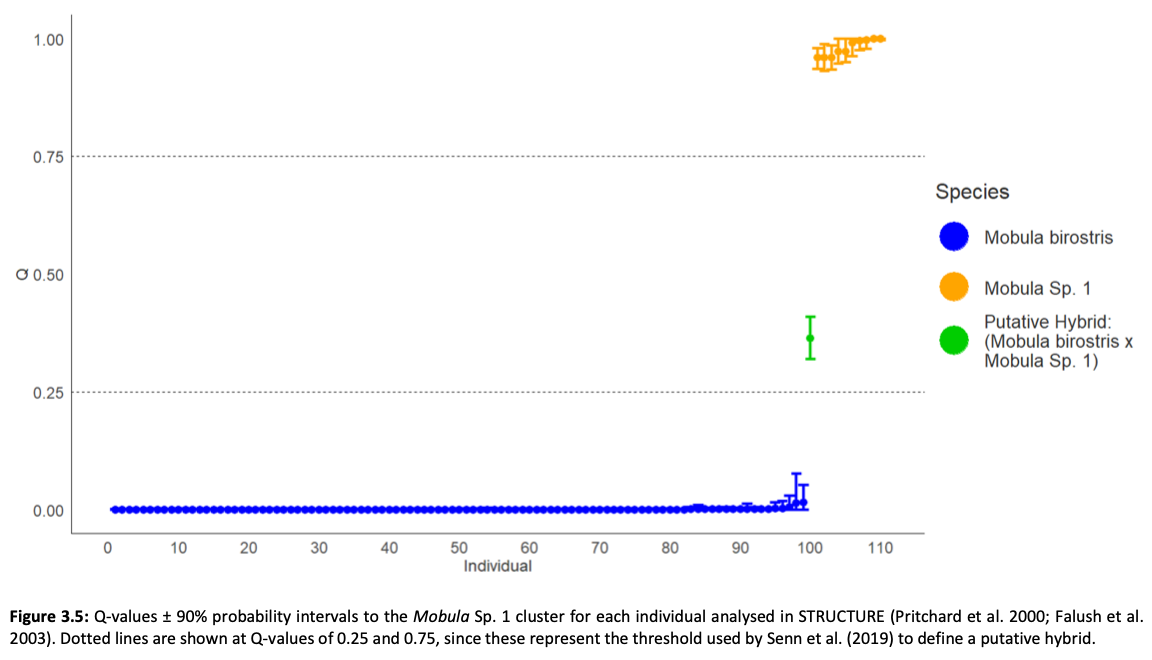
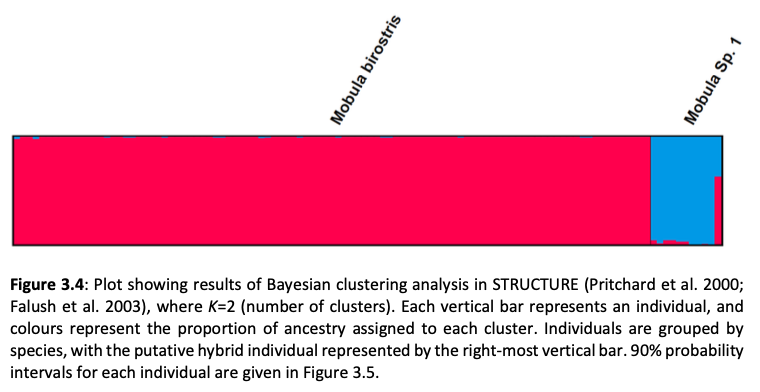
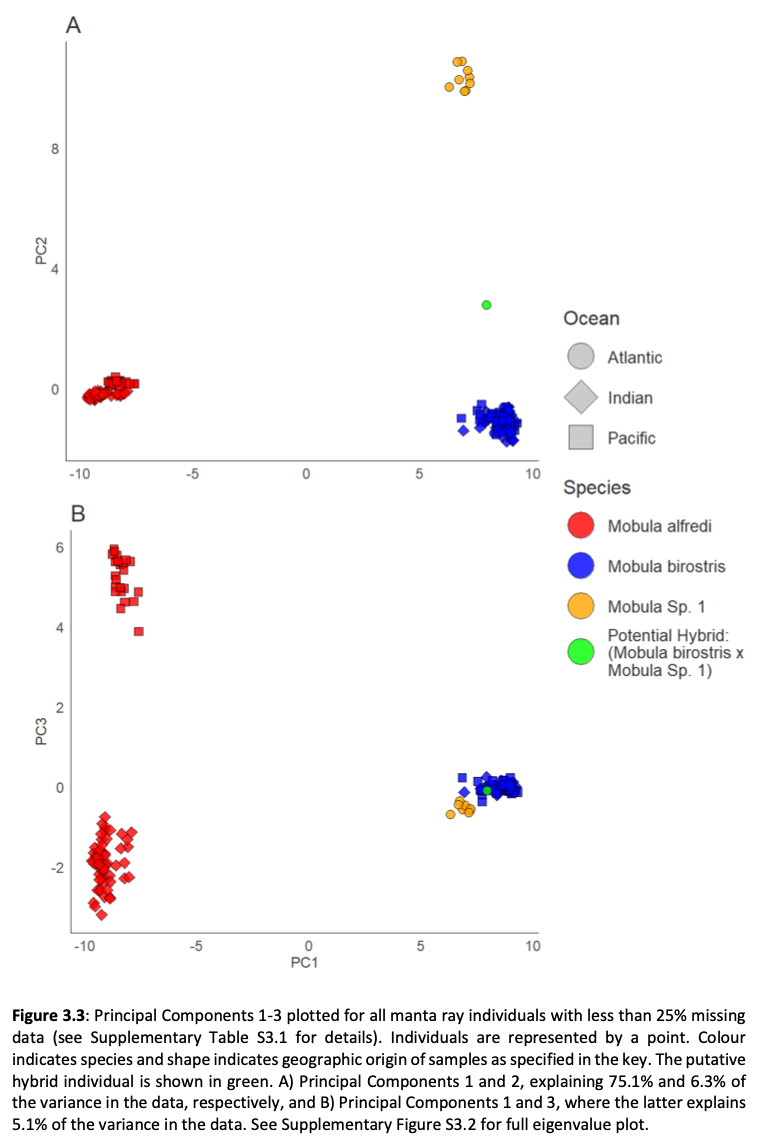
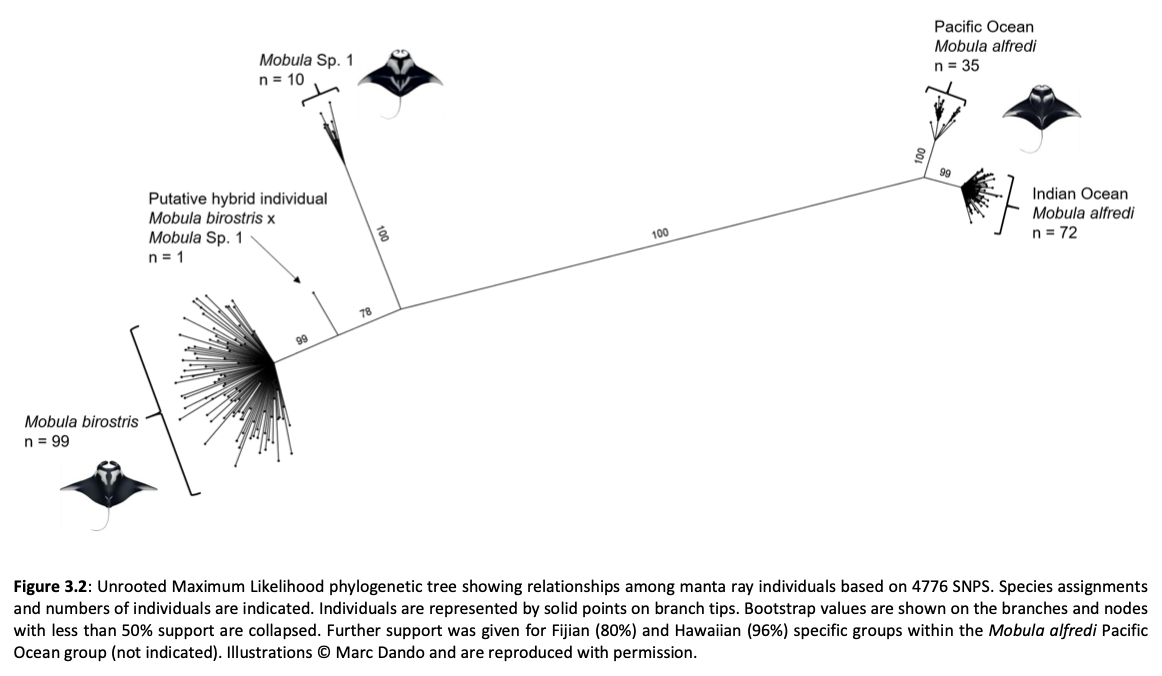
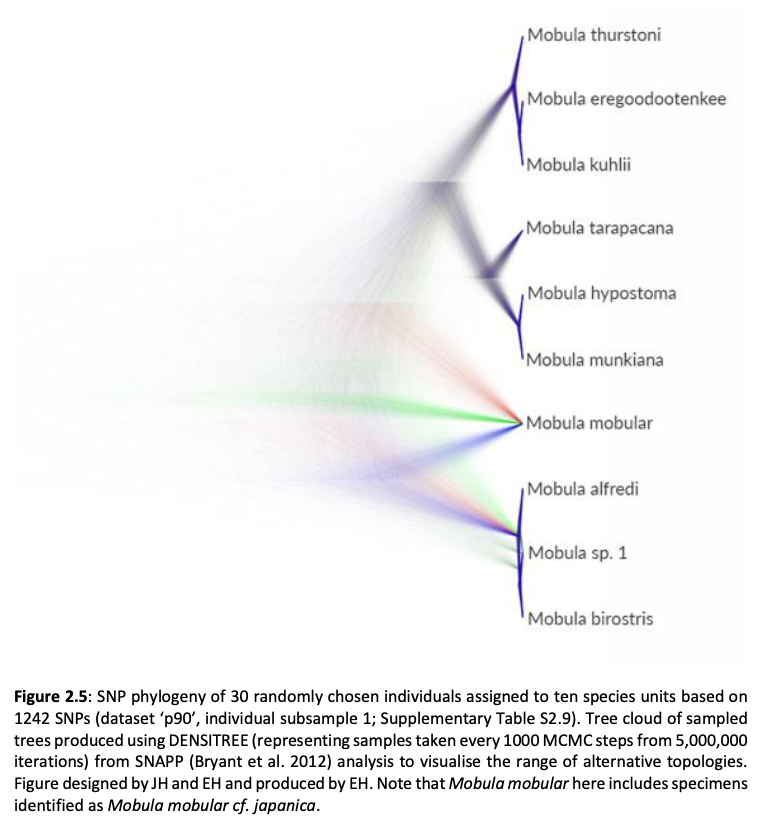
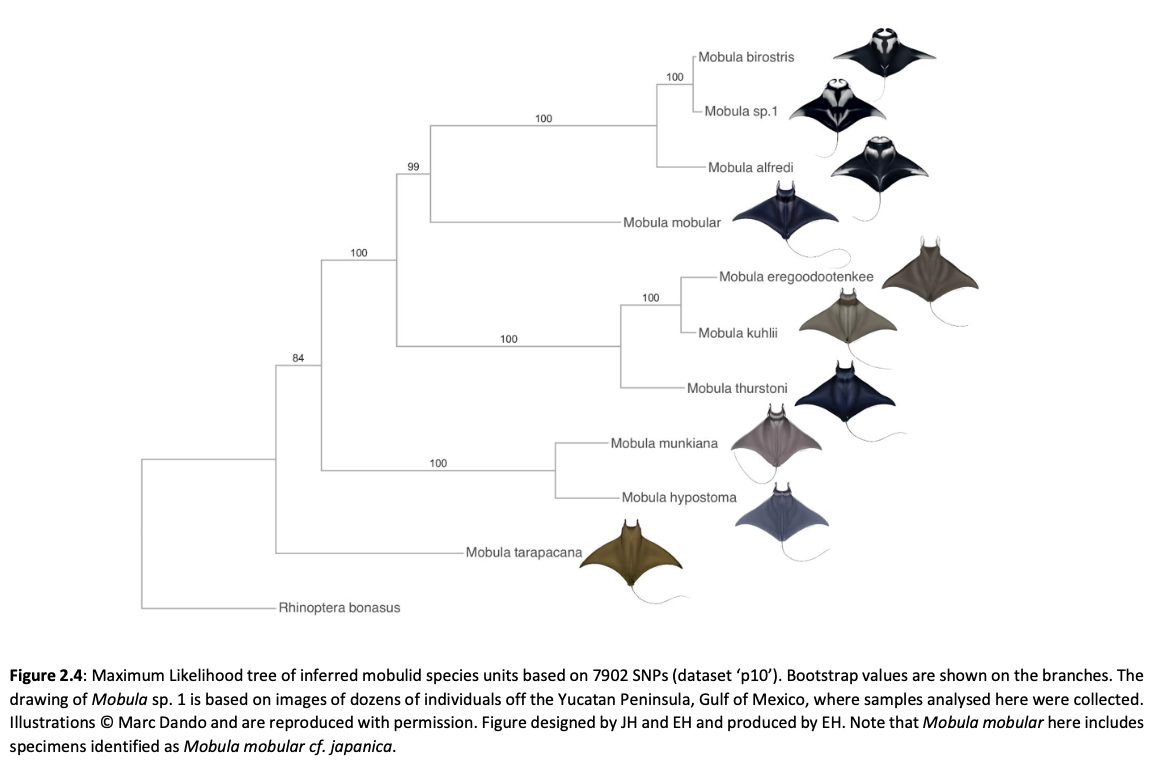
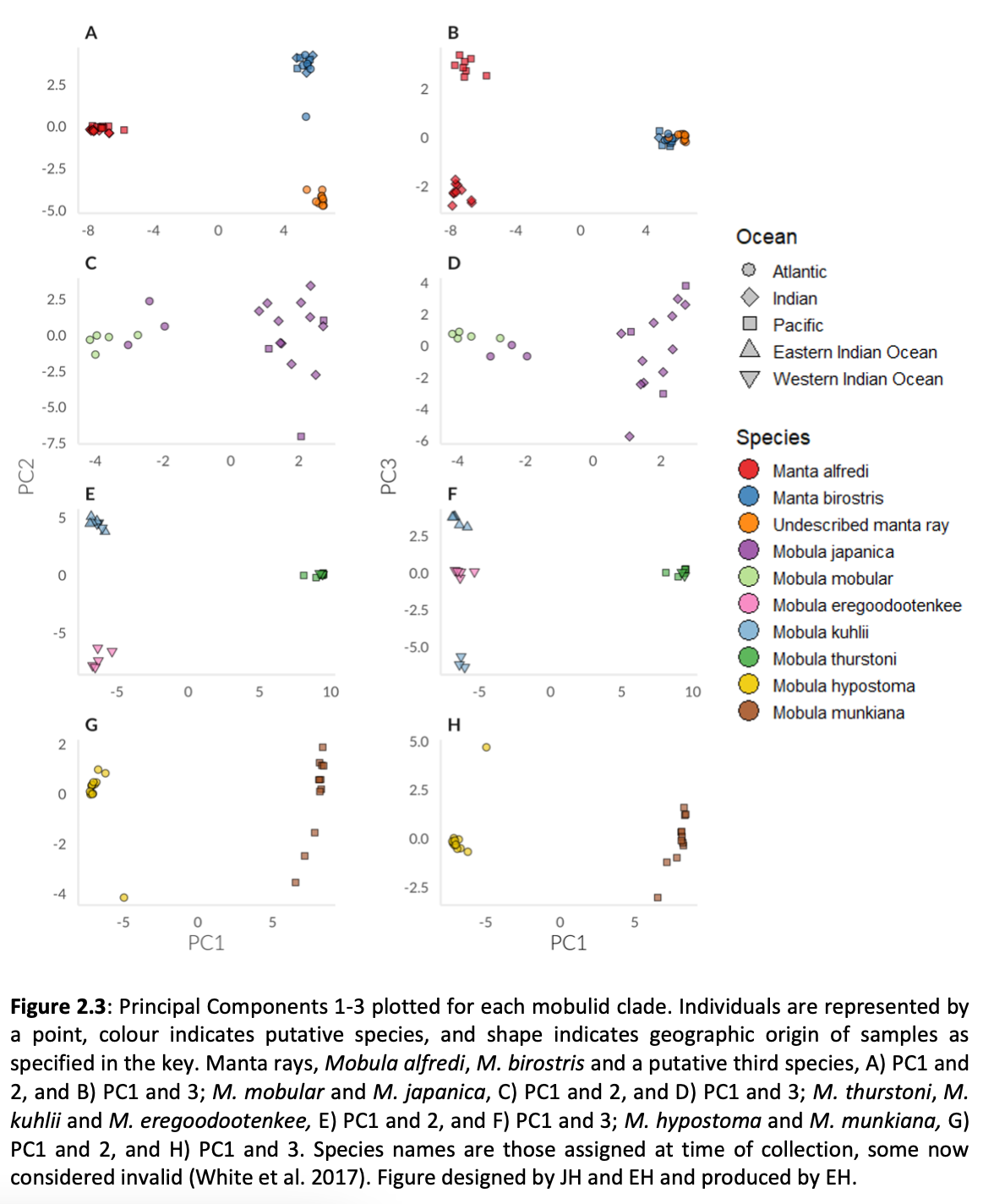
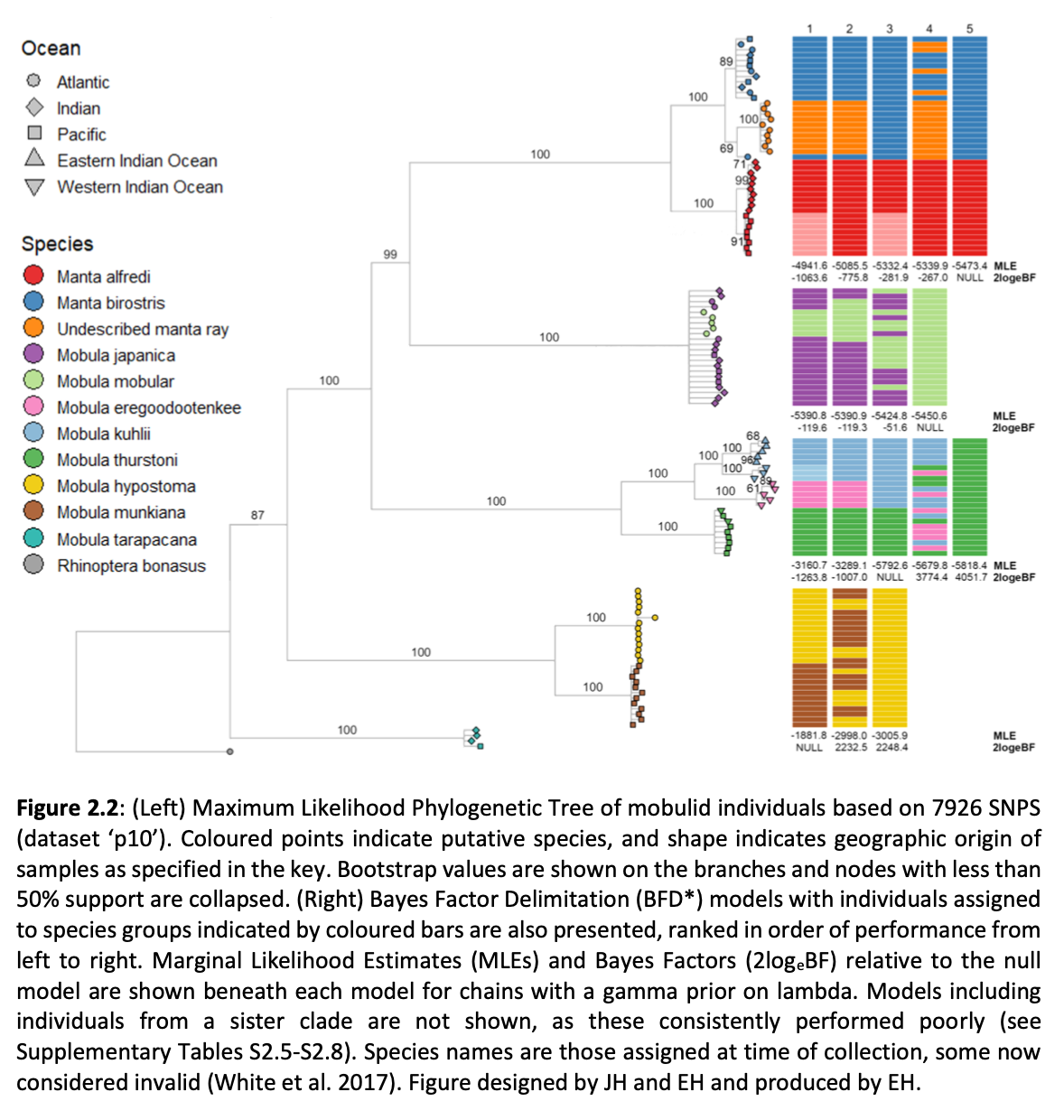
Here, genome-wide Single Nucleotide Polymorphism (SNP) data is generated from an exceptional global collection of mobulid tissue samples, representing all described species across their geographic ranges and is used to target these knowledge gaps. Phylogenomic reconstruction of the Mobulidae combined with species delimitation based on the multispecies coalescent identifies mismatches between currently recognised species, and species units optimal for conservation under international frameworks. Specifically, an undescribed species of manta ray is shown to be present in sympatry with the oceanic manta ray in the Gulf of Mexico, with some evidence of hybridisation. In addition, these data show two recently synonymised species to be distinct and reproductively isolated and reveals geographically mediated population structure in several species. Substantial incomplete lineage sorting is uncovered and standing variation in extinct ancestral populations is identified as a driver of phylogenetic uncertainty.
These data show that the lineage corresponding to the undescribed species of manta ray is associated with reduced genetic diversity, consistent with a pattern of peripatric speciation in isolation and highlighting conservation concerns for this species. Hybridisation between the oceanic manta ray, Mobula birostris, and the undescribed species of manta ray is confirmed for the first time but shown not to be associated with introgression. Such insights suggest that hybrids may be inviable, with conservation implications where unsuccessful reproductive investment in hybrid offspring is a concern in these species with slow life history traits. Extremely rapid and complete speciation in a marine system is presented, and highlights concerns associated with anthropogenic climate change and accompanying sea level changes on evolution in the oceans.
Population genetic structure is compared between two species of manta ray with contrasting habitat preferences. Whilst the reef manta ray (Mobula alfredi), shows a high degree of population structure among sampling locations, genome-wide SNPs indicate global genetic panmixia in the oceanic manta ray (Mobula birostris). Declining genetic diversity across the Pacific Ocean may be suggestive of successive founder events in the reef manta ray. Global genetic panmixia in oceanic manta rays may relate to past demographic processes, or differential dispersal among life stages. These highly contrasting patterns highlight the importance of evaluating population structure and adaptive divergence individually for related species of conservation concern, rather than relying on an assumption that closely related species display similar patterns. Collectively, our findings provide a substantial contribution to current knowledge pertaining to manta and devil rays.”
About The Author - Dr. Jane Hosegood
Whilst studying for an undergraduate degree in Zoology at the University of Sheffield, Jane was introduced to the importance of genetics when considering conservation and management strategies for wildlife populations, and decided that research in this area was something she was passionate about pursuing. She then went on to complete an MRes in Biodiversity and Conservation at the University of Leeds where she specialised in conservation genetics. It was whilst studying for her Masters that Jane was given the opportunity to join the Manta Trust Genetics Project and study for a PhD at the Molecular Ecology and Fisheries Genetics Laboratory at Bangor University. As part of this project, Jane generated a large genomic dataset for manta and devil rays and used this to investigate species boundaries and population structure at a global scale. Jane completed her PhD in 2019 and the results of this work will be invaluable for the future conservation of these species.
Read more about the MOBULID GENETICS PROJECT